-
Research
-
Publications
-
All publications
-
Benner, SA
-
Biondi, E
-
Bradley, K
-
Chen, C
-
Hoshika, S
-
Karalkar, N
-
Kim, HJ
-
Kim, MJ
-
Laos, R
-
Leal, NA
-
Li, Y
-
Richards, N
-
Shaw, RW
-
Spacek, J
-
Yang, ZY
-
People
-
Benner, Steven
-
Biondi, Elisa
-
Bradley, Kevin
-
Chen, Cen
-
Darling, April
-
Hoshika, Shuichi
-
Karalkar, Nilesh
-
Kim, Hyo-Joong
-
Kim, Myong-Jung
-
Laos, Roberto
-
Leal, Nicole
-
Li, Yubing
-
Richards, Nigel
-
Shaw, Ryan
-
Spacek, Jan
-
Yang, Zunyi
-
News and Events
-
Press Coverage
-
Our Foundation
|
The Origin of Life

The origin of life on Earth, Mars and elsewhere in the cosmos
Paleogenetic analyses have confirmed the historic role of RNA near the origin of life. However, many paradoxes have long made it seem unlikely that RNA could have emerged on a primitive Earth. First among these is the "tar paradox", the well-known propensity of organic molecules to devolve to give tar, absent Darwinian evolution. Another paradox remarks that RNA functions only in water, but is also slowly degraded in water.
Our group helped to introduce minerals in prebiotic chemistry to manage such paradoxes. Borate minerals were shown to help manage the tar paradox. An understanding of the history of bodies impacting Earth has helped resolve issues related to the oxidation-reduction potential of its atmosphere, and helped define the timing of the origin of life on Earth. A team at the Foundation recently showed that RNA is made from triphosphates on volcanic glass that was abundant at that time. Other researchers at FfAME have developed chemistry that might have yielded these triphosphates on early Earth. Similar chemistry is likely to have occurred on a young Mars. This research is now supported by the National Science Foundation.
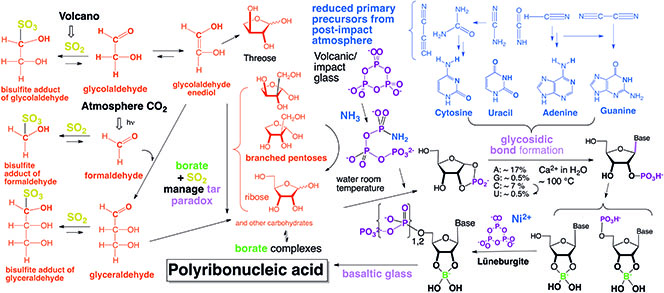
A relatively direct model connects simple organic molecules via minerals to the formation of prebiotic RNA.

The search for alien life
The Viking 1976 mission to Mars carried life detection tests that gave positive results. These results were discounted, however, because a gas chromatograph-mass spectometry experiment reported no organic molecules in the accessible Martian soil. Our group showed how these GC-MS results were misinterpreted. This, in turn, set the stage for current Mars missions.
Today, an affiliate of FfAME is building an agnostic life finder (ALF) to identify life in water on Mars, Europa, and elsewhere in the Solar System. This is based on our improving understanding of how life originated, as well as work in synthetic biology that defines the universe of possible genetic molecules. We are presently building an international team and planning the necessary steps to get ALF to Mars, on a private mission if necessary.
Our group's command of chemistry in geological contexts has allowed it to develop the biology-relevant chemistry of other astrobiology targets in the Solar System. These include Titan, where the we asked whether life might be sustained in truly "weird" environments, such as its methane oceans. This led to NASA's latest flagship mission to fly to this moon of Saturn. A team led by FfAME researchers has made major progress in understanding the organic chemistry in the Venusian clouds. Research made in our group informs the first private interplanetary mission organized by MIT Venus Cloud Life group and sponsored by Rocket Lab. The mission attempts to detect fluorescence provided by organic molecules in the clouds of Venus in 2023.
Our teams have addressed possible biosignatures on many of the worlds in our Solar System, from left to right, Venus, Mars, Europa, and Titan.
|
|
|



